© LBDV / S.Peron
In response to injury, some animals can activate a repair process, which restores body integrity and functionality. Repair can be limited to healing of the damage, or lead to the regeneration of missing structures, including large portions of the body. The mechanisms governing the switch from wound healing to regeneration are still largely unknown across animals, and likely involve mechanical and transcriptional inputs. Clytia offers a unique possibility for disentangling the relative contributions of mechanical and transcriptional cues in the activation of the regenerative program. We could show that this jellyfish displays excellent regenerative capacities, and that the fate of an injury depends on the interplay between mechanical forces, notably actomyosin contractility, and Wnt signalling. We are characterizing and modelling how all the different parameters – mechanical forces, transcriptional regulation and tissue polarity – are integrated at the cellular and tissue levels, and translated into a system-level regenerative program. This multiscale research program combines omics (scRNAseq and ATACseq), live imaging of transgenic reporters, forces quantification and manipulation, and modelling.
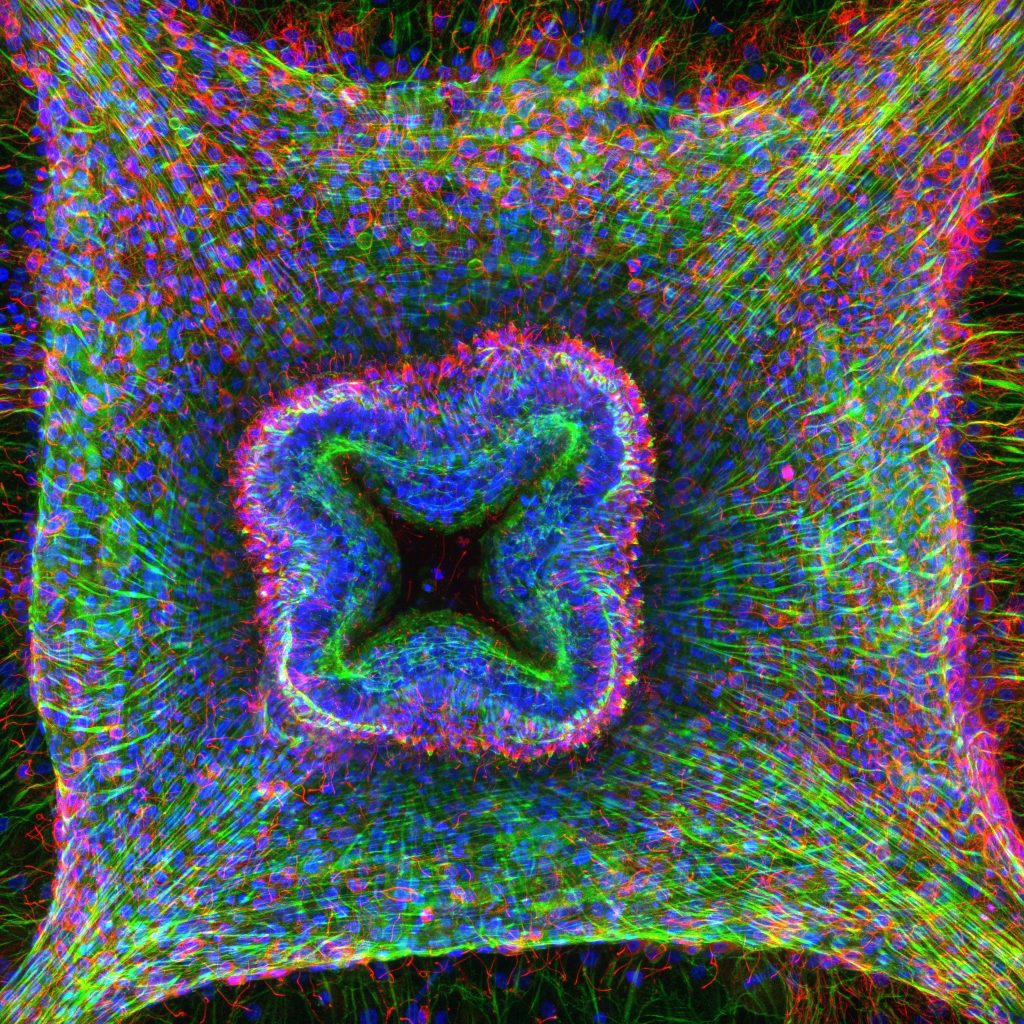
© LBDV / S.Peron
Jellyfish swim in the water column thanks to striated muscles located on the underside of their umbrella. These striated muscles strongly resemble the striated muscles typical of bilaterian animals, despite the fact that they likely evolved independently. Combining live microscopy, single cell transcriptomics, super-resolution immunofluorescence and functional approaches, we are characterizing the molecular makeup of jellyfish striated muscles, addressing how they develop, function and have evolved, using both the hydrozoan Clytia hemisphaerica and the scyphozoan Pelagia noctiluca.
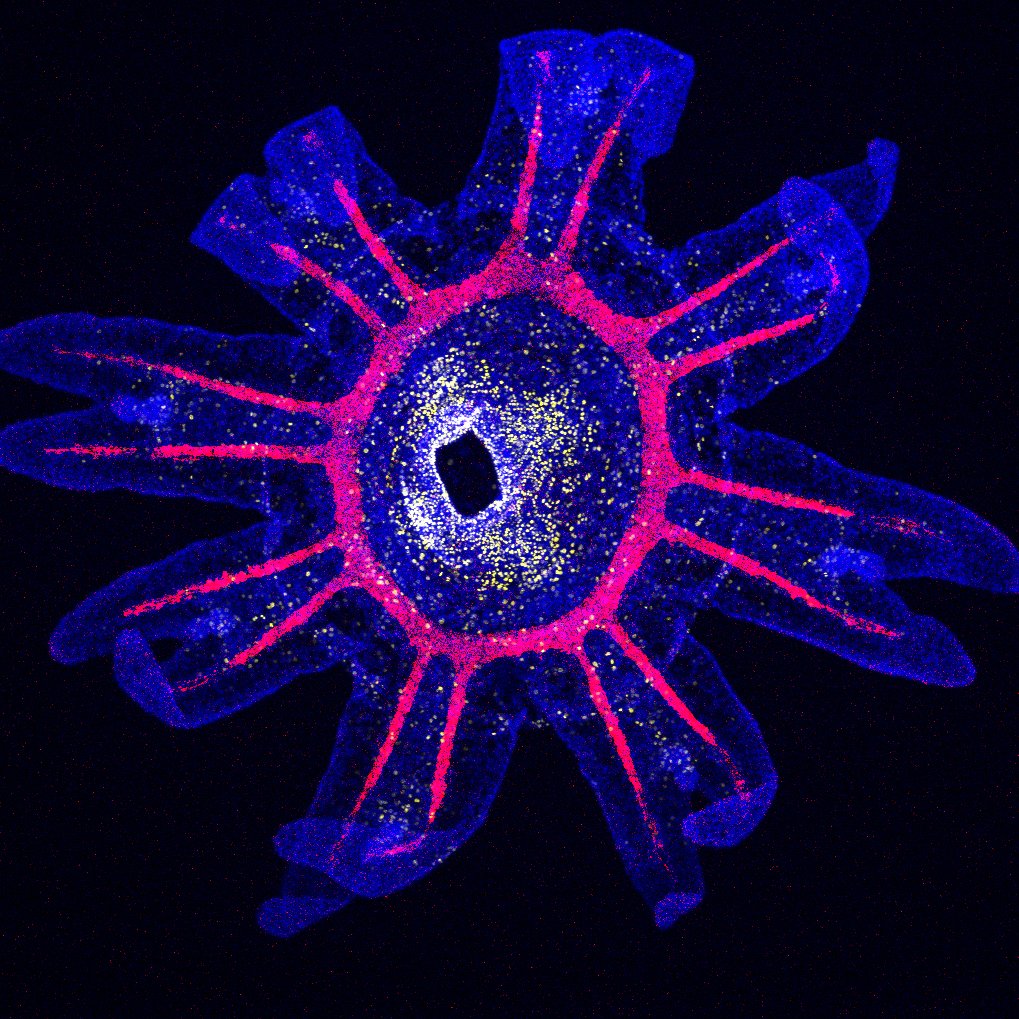
© LBDV / B.Salmon
Medusozoan cnidarians display some of the most complex metazoan life cycles. The canonical medusozoan life cycle alternates between a swimming planula larva, a benthic polyp and asexually-produced pelagic medusae. Many medusozoan clades have lost either the polyp or the medusa form, dramatically shifting the species ecology towards the benthic or the planktonic environment. These are major life history transitions, and our current understanding of their genomic, ecological and developmental implications is very limited. In order to start addressing this major knowledge gap, we are combining genomic, transcriptomic and phylogenetic approaches on multiple medusozoan species. We are currently focusing on two clades, the Leptothecata (where multiple loss of the medusa stage occurred) and the Pelagiidae (showing loss of the polyp stage in the Pelagia lineage).

© LBDV / A.Jan
Funding









